SourceData-NLP
Integrating curation into scientific publishing to train AI models
The largest Named Entity Recognition (NER) and Named Entity Linking (NEL) dataset in biomedical sciences. Integrating AI-ready curation directly into the publishing workflow at EMBO Press.
Also available in:
🇪🇸Leer en EspañolSourceData-NLP: When Publishing Meets AI
What if we could capture structured knowledge from scientific papers the moment they're published, rather than mining them years later? That's the revolutionary idea behind SourceData-NLP—the largest Named Entity Recognition (NER) and Named Entity Linking (NEL) dataset in biomedical sciences to date. The paper is approved for publication in the Oxford University Press journal Bioinformatics.
The Vision: Publishing as Knowledge Infrastructure
Scientific publishing has been humanity's primary mechanism for sharing knowledge for centuries. Yet this rich repository remains locked in unstructured text, requiring enormous effort to extract and organize. SourceData-NLP reimagines this process by embedding AI-ready curation directly into the publishing workflow at EMBO Press.
Rather than retrospectively annotating papers, we work with authors during peer review to capture structured data as part of the publication process itself. This approach transforms scientific publishing from a static archive into a living, machine-readable knowledge infrastructure.
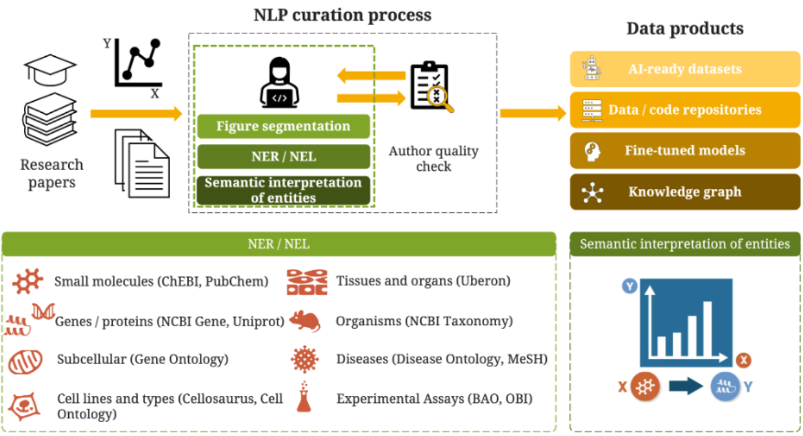
Figure 1: The SourceData-NLP curation workflow. The integration of NLP-assisted curation into the publishing process enables the capture of structured knowledge from research papers. Authors validate entity annotations during peer review, ensuring high-quality training data. The process generates multiple data products including machine-learning ready datasets, fine-tuned models, and semantic knowledge graphs connecting biomedical entities across the literature.
By the Numbers
SourceData-NLP is unprecedented in scale and scope:
- 661,862 annotated entities across 8 biomedical categories
- 623,162 entities linked to external ontologies and databases
- 62,543 annotated figure panels from 18,689 scientific figures
- 3,223 research articles from 25 leading journals in molecular and cell biology
But size isn't everything. What makes SourceData-NLP unique is its focus on figure legends—the narrative core of scientific evidence where experimental results are described. Unlike datasets built from isolated sentences, SourceData-NLP preserves the rich context of real scientific communication.
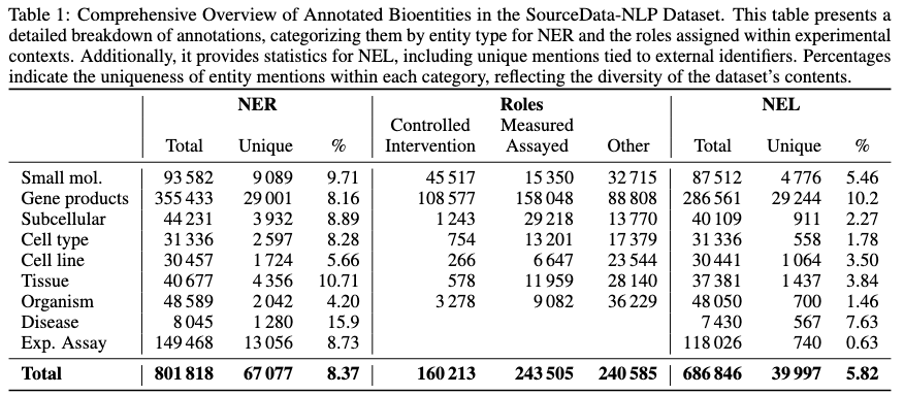
Table 1: Comprehensive overview of annotated bioentities in the SourceData-NLP dataset. The table presents detailed statistics for Named Entity Recognition (NER), experimental role assignments, and Named Entity Linking (NEL) across all entity categories. The uniqueness percentages reflect the diversity of entity mentions, with experimental assays showing the lowest uniqueness (0.63%) due to repeated methodologies, while small molecules show the highest (9.71%) reflecting diverse chemical compounds studied across papers.
What We Annotate
The dataset captures the essential elements of biomedical research:
Eight biological entity types: small molecules, gene products, subcellular components, cell lines, cell types, tissues, organisms, and diseases
Experimental context: Beyond simply identifying entities, we annotate their roles in experimental designs and the methodologies used to study them
Semantic relationships: Each entity is linked to authoritative databases, enabling disambiguation and connecting findings across the literature
This granular annotation, validated by original authors, ensures the highest quality training data for AI models.
Why This Matters
Integrating curation into publishing isn't just about building a better dataset—it's about fundamentally rethinking how scientific knowledge flows. When structured data is captured at the source, it becomes immediately available for:
Knowledge Discovery
AI models can identify patterns and relationships across millions of experiments that no human could manually track.
Accelerated Research
Researchers can instantly query what's known about specific genes, diseases, or experimental approaches without reading thousands of papers.
Reproducibility & Rigor
Standardized entity annotation during publishing helps resolve terminological ambiguities and unclear concepts before they propagate through the literature.
AI Training at Scale
Large language models trained on SourceData-NLP understand not just biomedical terminology, but the experimental context in which discoveries are made.
Downstream Applications
The SourceData-NLP dataset enables a new generation of biomedical AI tools:
Literature Intelligence
- Automated systematic reviews and meta-analyses
- Real-time research trend tracking and gap analysis
- Evidence synthesis across decades of publications
Drug Discovery
- Novel target identification through entity relationship mining
- Drug repurposing by connecting known compounds to new disease contexts
- Adverse effect prediction from experimental literature
Clinical Translation
- Linking basic research findings to clinical applications
- Building comprehensive disease-gene-drug knowledge graphs
- Supporting precision medicine with literature-derived evidence
Research Assistance
- Experimental design recommendations based on similar studies
- Methodology suggestions for specific entity types
- Hypothesis generation from unexpected entity co-occurrences
Scientific Understanding
- Multimodal figure interpretation combining images and captions
- Automated extraction of experimental protocols and conditions
- Tracking the evolution of scientific concepts over time
The Path Forward
SourceData-NLP demonstrates that publication-integrated curation is not only feasible but transformative. As the dataset continues to grow with each new publication at EMBO Press, it becomes an increasingly powerful resource for training AI models that understand the nuances of biomedical research.
This is more than a dataset—it's a blueprint for how scientific publishing can evolve to serve both human readers and AI systems, accelerating the pace of discovery while maintaining the rigor that defines quality science.
Tools & Technologies
The SourceData-NLP project leverages a comprehensive stack of modern AI and data technologies:
- HuggingFace Transformers - Pre-trained models and fine-tuning infrastructure
- PyTorch - Deep learning framework for model training and inference
- Python - Primary programming language for data processing and model development
- GPU Computing - Accelerated training and inference for large language models
- NLP - Natural Language Processing pipelines for entity recognition and linking
- Neo4j - Graph database for knowledge graph storage and querying
- Graph Database - Storage and retrieval of semantic relationships between entities
Access the Dataset
- Paper: arXiv:2310.20440
- EMBO on HuggingFace: HuggingFace - EMBO
- Dataset: HuggingFace - EMBO/SourceData
- Model Repository: GitHub - soda-model
- Data Repository: GitHub - soda-data
Citation
@article{AbreuVicente2023,
title={The SourceData-NLP dataset: integrating curation into scientific publishing for training large language models},
author={Abreu-Vicente, Jorge and Sonntag, Hannah and Eidens, Thomas and Lemberger, Thomas},
journal={arXiv preprint arXiv:2310.20440},
year={2023},
month={October}
}
By capturing knowledge at its source—the moment of publication—we're building the foundation for AI systems that can truly understand and advance biomedical research.
